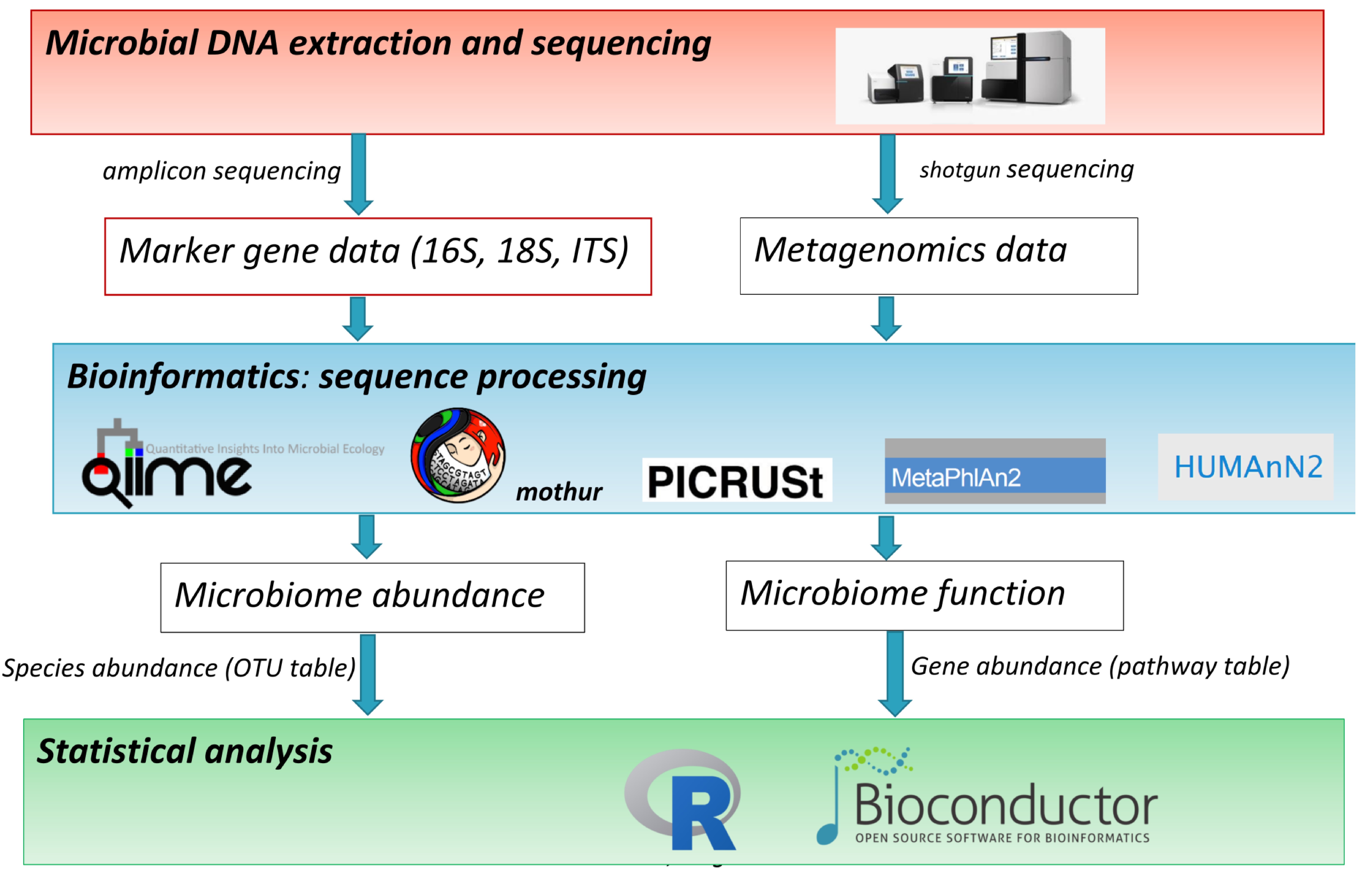
Overall marker gene abundance and redundancy for metagenomes from Kelp... | Download Scientific Diagram
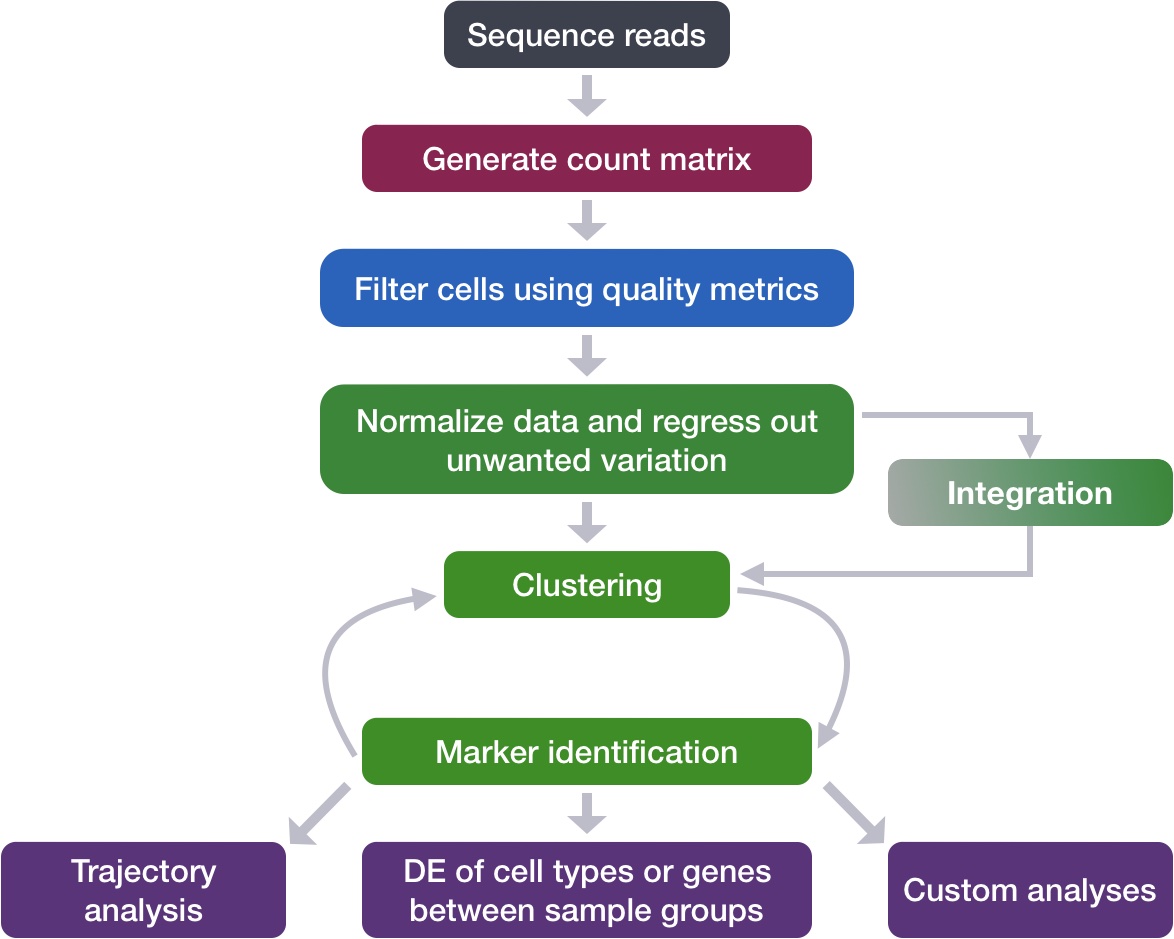
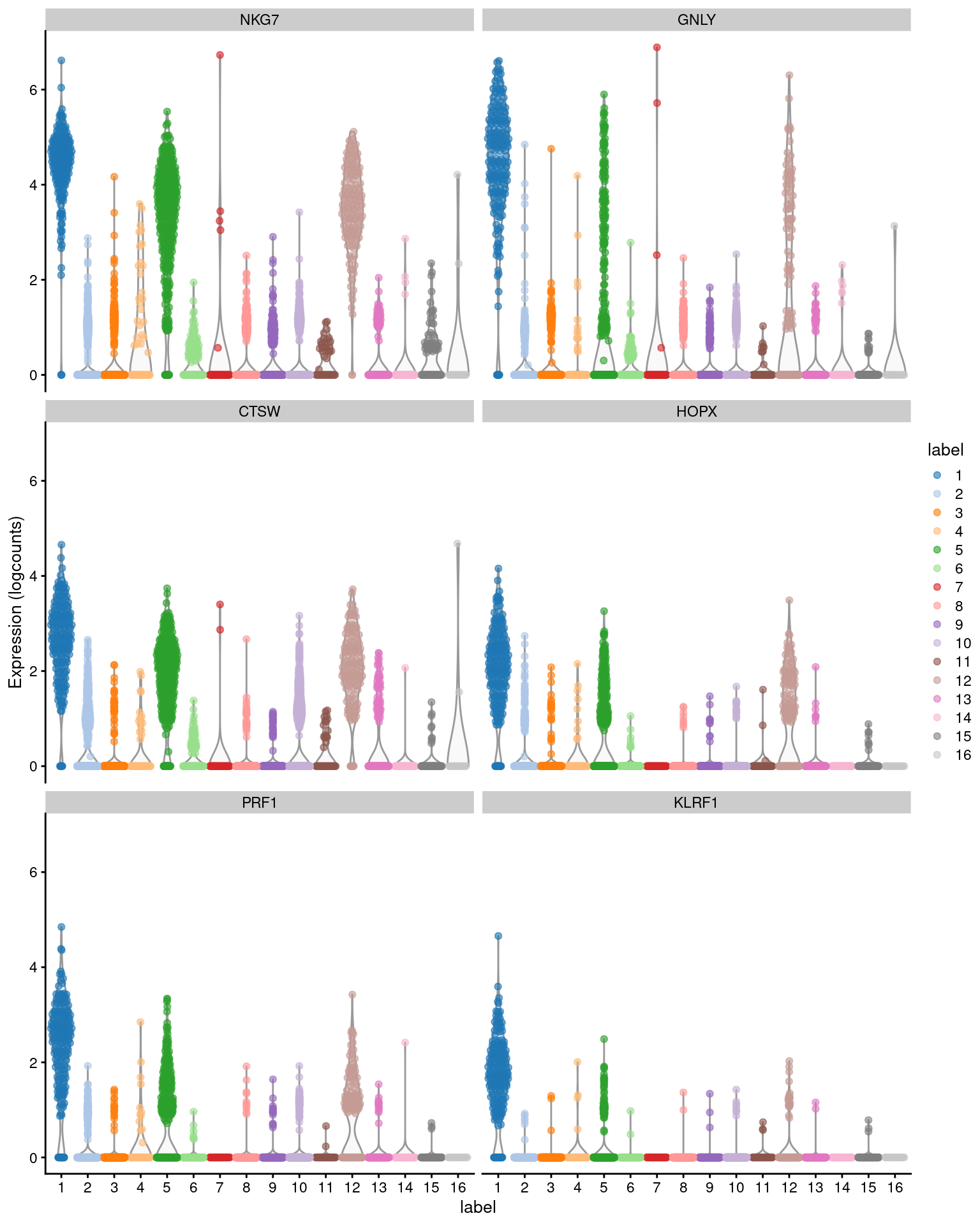
Single-cell RNA-seq: Normalization, identification of most variable genes | Introduction to single-cell RNA-seq
Simultaneous Quantification of Multiple Bacteria by the BactoChip Microarray Designed to Target Species-Specific Marker Genes | PLOS ONE

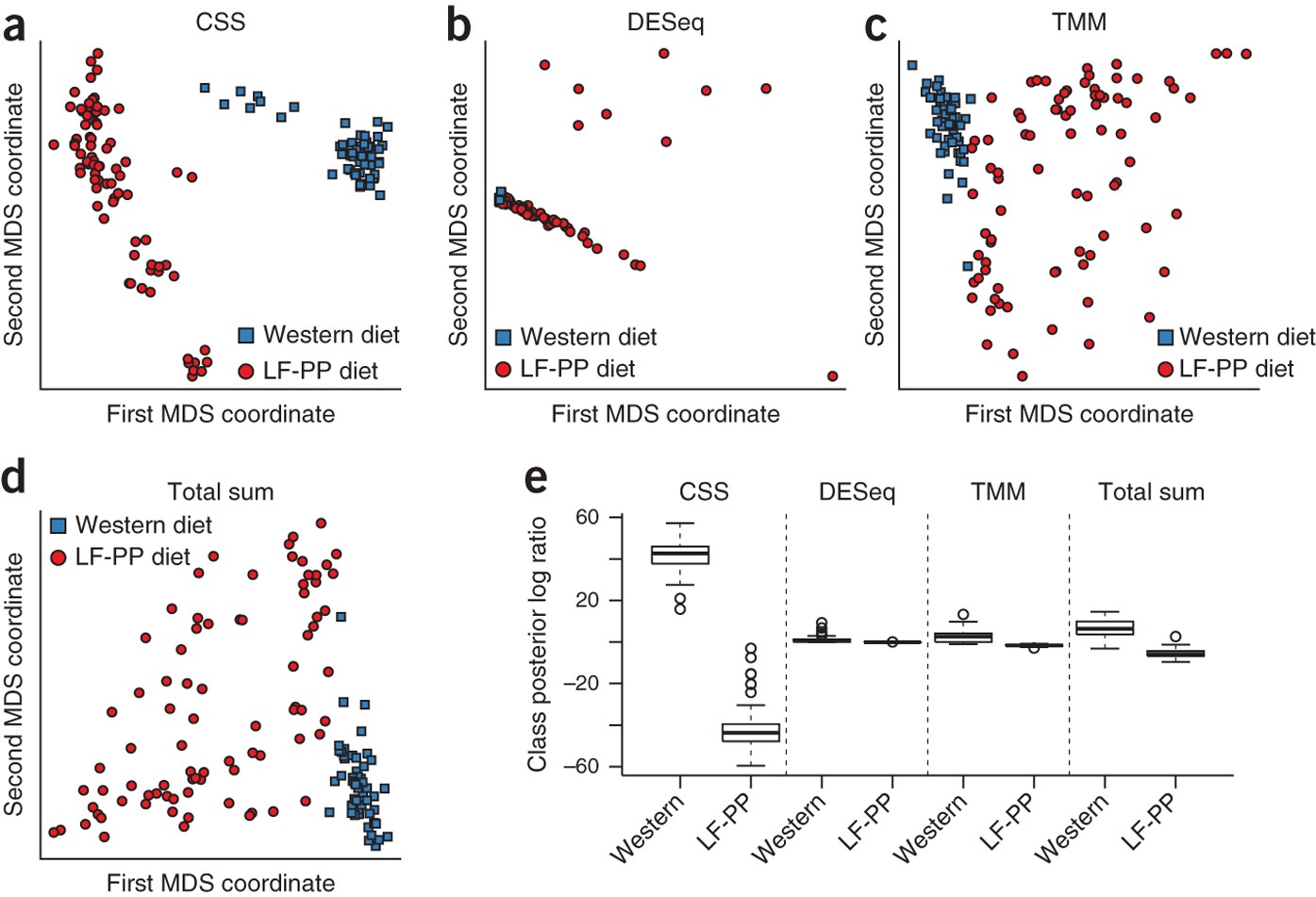
A neural network‐based framework to understand the type 2 diabetes‐related alteration of the human gut microbiome - Guo - 2022 - iMeta - Wiley Online Library
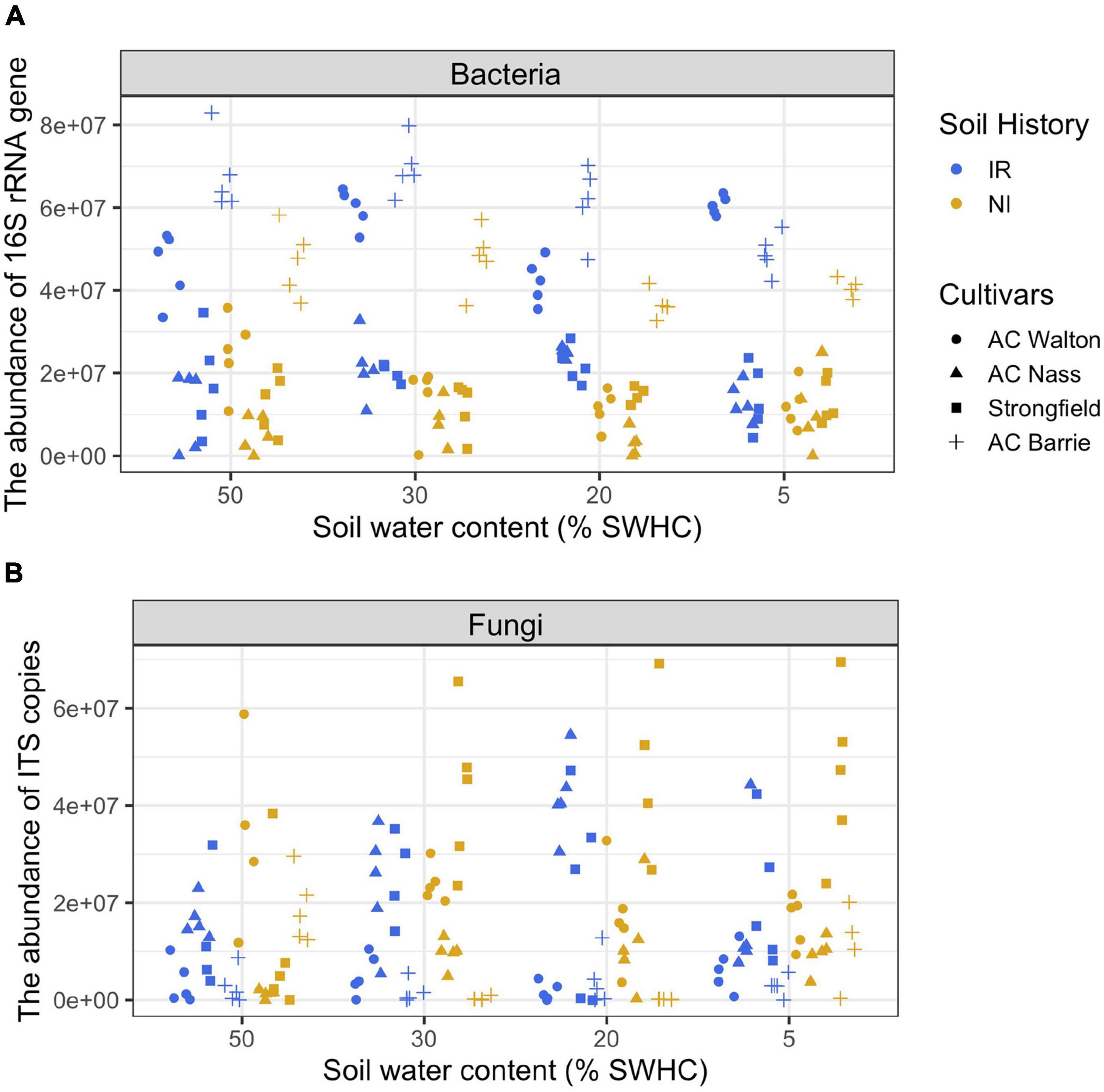
Frontiers | Relative and Quantitative Rhizosphere Microbiome Profiling Results in Distinct Abundance Patterns

Relative transcript abundances of the wounding/jasmonic acid marker... | Download Scientific Diagram

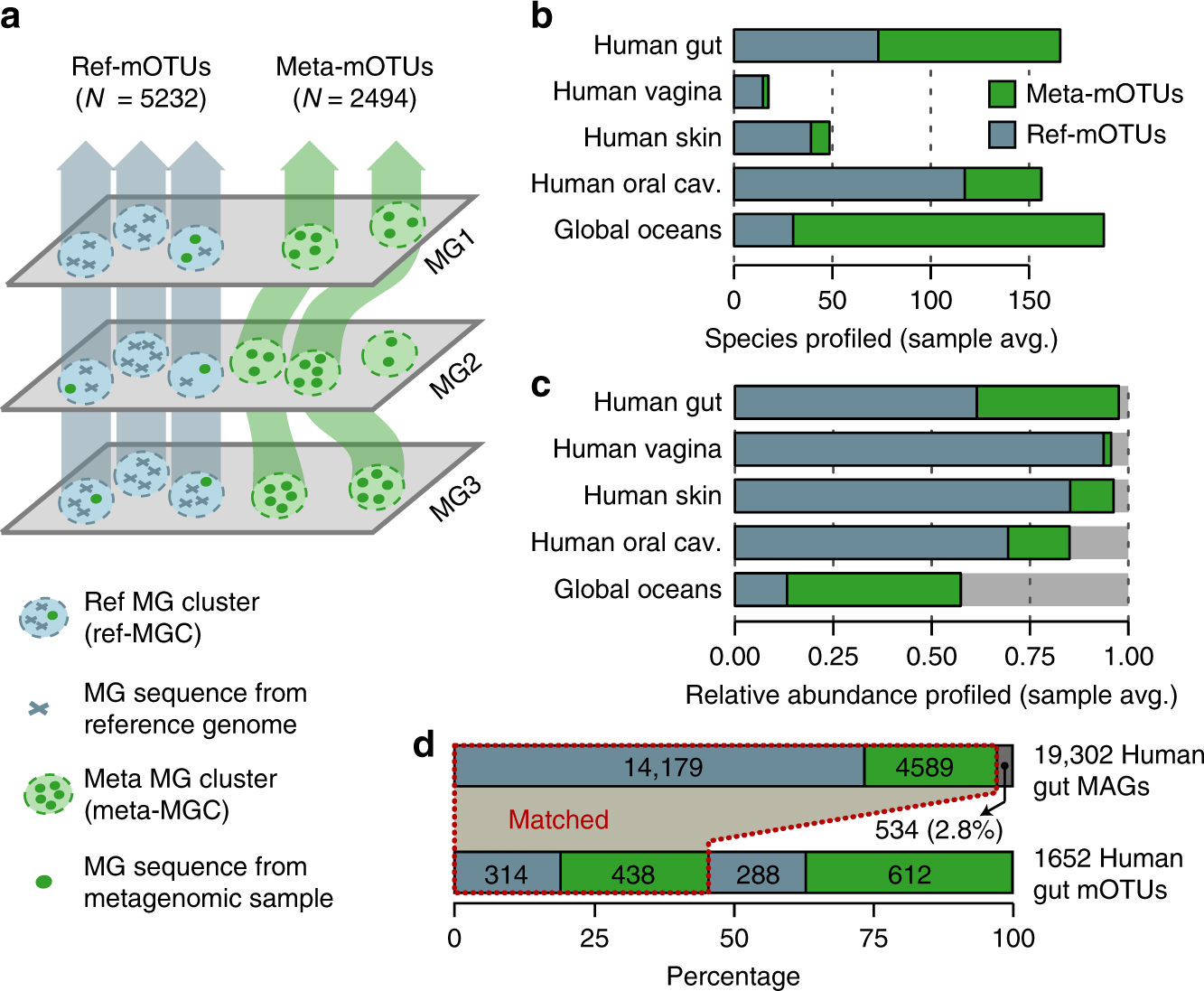
Overview of bacterial abundances inferred using 16S rRNA marker gene... | Download Scientific Diagram
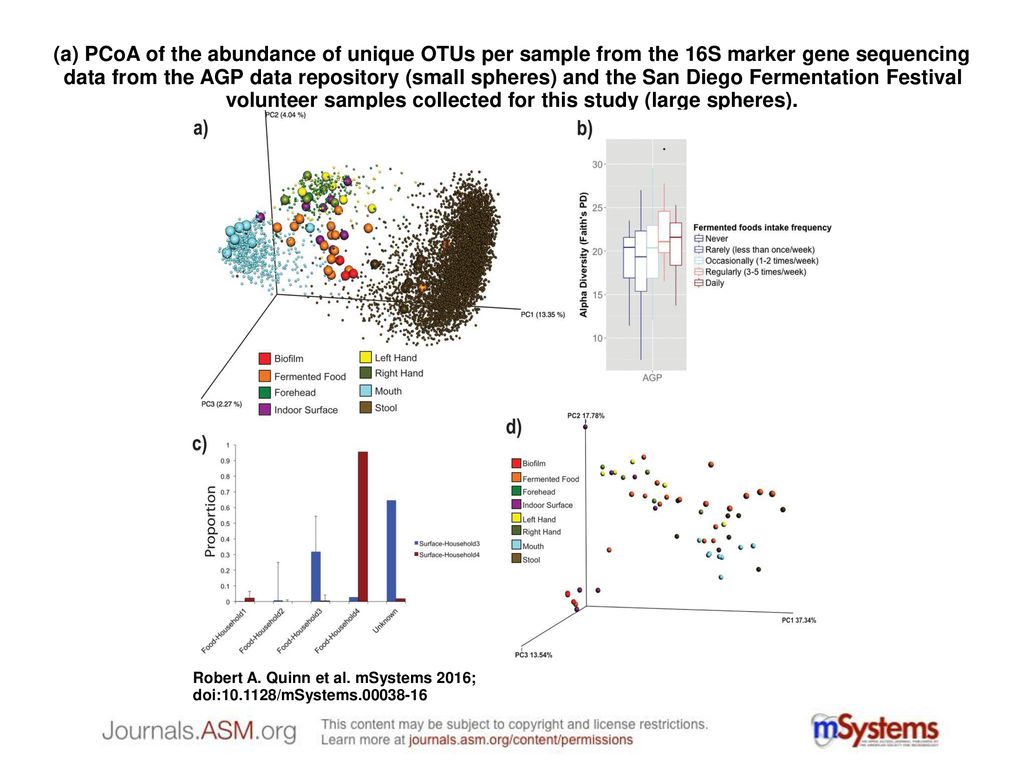
a) PCoA of the abundance of unique OTUs per sample from the 16S marker gene sequencing data from the AGP data repository (small spheres) and the San Diego. - ppt download

Inferring gene regulation from stochastic transcriptional variation across single cells at steady state | PNAS

Single-Nucleus RNA-Seq Is Not Suitable for Detection of Microglial Activation Genes in Humans - ScienceDirect

Distribution of fungi (≥1% abundance) based on the ITS2 marker gene... | Download Scientific Diagram